COVID19 protein-protein wars: designing agents to block infection
It is inspiring to see people on all levels come together to join the fight to design treatments and vaccines against COVID-19. From major Pharma, funding agencies (eg The Bill and Melinda Gates foundation, and NIH), scientific institutions, and taskforces, to crowdsourcing and citizen science. Here is a recent analysis of the current state of research and development.
This prompted me to share some thoughts and ideas regarding how protein engineering and structural biology can play an important role in the design and production of protein therapeutics that could be effective against SARS-CoV-2.
In the previous post I ended with a quick mention of the structure of the virus spike receptor binding domain (RBD)-Fab antibody complex by Ian Wilsons group at Scripps. Since then the paper describing that work has been reported. The Fab, derived from the antibody CR3022 originally isolated during the 2003 SARS outbreak, cross-reacts with SARS-CoV-2. Although CR3022 doesn't compete for ACE2 binding and is not neutralizing, the RBD-CR3022 structure shows a highly conserved RBD site on SARS-CoV and SARS-CoV-2. The structure also reveals why CR3022 binds SARS-CoV-2 less tightly than SARS-CoV, raising the exciting possibility of using protein engineering to produce a neutralizing antibody that might confer in vivo protection. As the authors point out, this is a fantastic example of how structural data will inform design of antibodies for passive immune protection as well as vaccine design.
See all the protein-protein interactions of SARS-CoV-2 at the EMBL-EBI complex portal.
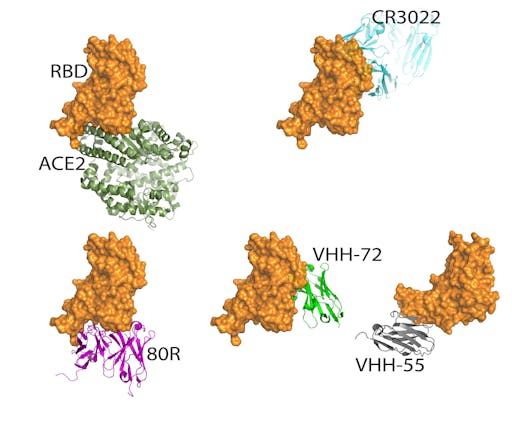
So what are the challenges in obtaining, isolating or designing antibodies to block the interaction between the RBD and the ACE2 receptor?
The concave shape of the cleft on the RBD where it docks with ACE2 presents a challenge for antibodies, which prefer flat or convex surfaces. SARS-CoV-2 uses the same host receptor (ACE2) as SARS-CoV – since the SARS outbreak in 2003 several neutralizing antibodies that bind the RBD of SARS-CoV have been structurally characterized. In fact, many of these compete with the ACE2-binding site. One of them, the neutralising antibody 80R, overlaps entirely with the ACE2 binding site on the RBD of SARS-Co-V, showing that in this case it is possible for antibodies to block ACE2 binding despite the concave cleft on the RBD. Given that in the past 15 years the interactions between RBDs of several highly related coronaviruses and host receptors have been structurally characterised, and the high degree of sequence and structural homology between SARS-CoV and SARS-CoV-2, it is likely that antibodies that offer passive immune protection for SARS-CoV-2 are on the horizon.
The concave shape of the cleft on the RBD where it docks with ACE2 presents a challenge for antibodies, which prefer flat or convex surfaces. SARS-CoV-2 uses the same host receptor (ACE2) as SARS-CoV – since the SARS outbreak in 2003 several neutralizing antibodies that bind the RBD of SARS-CoV have been structurally characterized. In fact, many of these compete with the ACE2-binding site. One of them, the neutralising antibody 80R, overlaps entirely with the ACE2 binding site on the RBD of SARS-Co-V, showing that in this case it is possible for antibodies to block ACE2 binding despite the concave cleft on the RBD. Given that in the past 15 years the interactions between RBDs of several highly related coronaviruses and host receptors have been structurally characterised, and the high degree of sequence and structural homology between SARS-CoV and SARS-CoV-2, it is likely that antibodies that offer passive immune protection for SARS-CoV-2 are on the horizon.
Size isn’t everything: antibody fragments
There are several antibody fragments that are smaller and thus more able to fit into the concave ACE2-binding cleft on the RBD. These are typically referred to single-domain antibodies or nanobodies. Examples include fully human single variable domains (VH), camelids (VHH), and shark nanobodies (V domains of immunoglobulin new antigen receptor (vNAR).
Single domain antibodies offer several advantages over their full size counterparts, including high stability and aggregation-resistance, reducing downstream manufacturing complexity and costs.
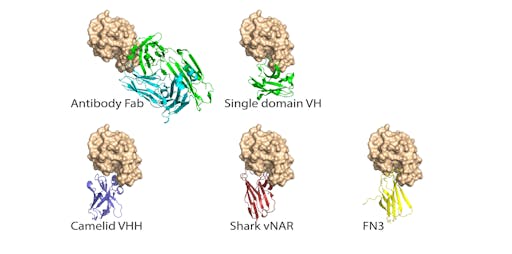
Work is already underway looking at how nanobodies might be useful as COVID19 therapeutics: a recent report showed that two single domain camelid antibodies (VHH-72 and VHH-55) bind MERS-CoV RBD and SARS-CoV RBD and exhibit cross-reactivity against the SARS-CoV-2 RBD. The authors have just deposited the crystal structures of both RBD-VHH complexes: PDB entry for VHH-72 is here (VHH-55; PDB 6WAR is here).
Alternatives to antibodies: the non-antibody scaffold zoo
There are now more than 12 classes of uniquely-shaped proteins that offer alternatives to antibodies and, but perform similar functions, particularly where high-affinity binding to a target is required. One particlar type of non-antibody scaffold, or monobody, is the FN3 domain. These have the advantage of being small, very stable, do not have disulphide bonds, and are easy and cheap to make in large quantities using bacterial expression.
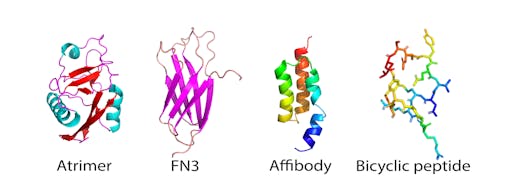
We recently engineered a hyperstable version of the FN3 domain that is robust to protein engineering - I will use this here to explore whether it, or a similar class of small protein scaffold could offer any advantages for the development of agents that interact with the SARS-CoV-2 RBD.
Very simple placement of the FN3 domain into the cleft normally occupied by ACE2 shows that it is, remarkably, about the right size, and shape for its convex binding site to fit snugly inside the concave cleft of the SARS-CoV-2 RBD. Nothing elaborate has been done here – no computational docking or calculations, just simple, manual docking by eye. But it is enough to see shape complementarity to warrant further interest. Analysis of the ACE2-RBD interface reveals a snug fit except for at the centre, where this is a hole – there are no interactions between the RBD and ACE2 here. This may represent an opportunity for design using such small, penetrating surfaces offered by monobodies.
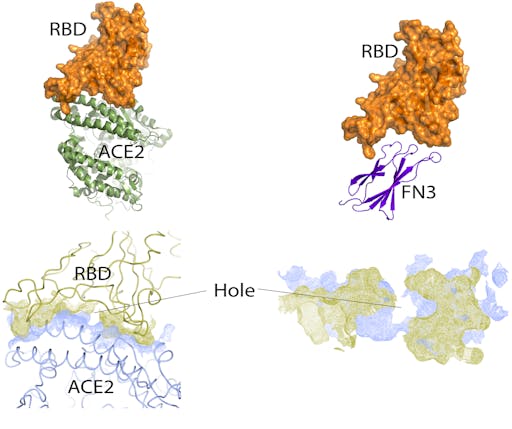
As of writing, I cannot find any reports investigating non-antibody scaffolds as potential protein therapeutics that target SARS-CoV or SARS-CoV-2.
OK, this is an oversimplification - where would you go from here? As with antibodies, the typical route would be to perform laboratory evolution, that is, to generate large libraries of variants where the sequences of the binding loops are randomized, and select for binding to RBD. It is important to point out that this certainly does not guarantee that these molecules will bind in the right place - it will be important to generate a panel of binders and select those that compete with ACE2 binding. But – if one is found that does bind in the RBD cleft, their ease and low cost of production, high thermal and colloidal stability (for example they may not require cold-chain storage), allowing the possibility of use in nebulizers or inhalers, might prove to be a competitive advantage over antibodies.
There are many obstacles still to overcome. The first of these is circulating plasma half-life (antibodies solve this using their large molecular weight and their Fc region, plus glycosylation). There are several examples of attaching large molecules such as PEG to FN3 however to achieve this, as well as other such fusions, for example with Fc regions and even mono- and multi-specific antibodies (offering solutions to the VH/VL mis-pairing problem). The second hurdle is unwanted immunogenicity. Again, this is not unsurmountable – it is possible to deimmunize the protein sequence by removing T cell epitopes identified either in silico or using T cell stimulation assays.
Should such obstacles be overcome, the high stability of monobodies and nanobodies means other routes of administration such as aerosols, which could be particularly attractive for respiratory diseases such as COVID19, become more practical. Thermal stability solves the need for cold-chain storage, a problem in resource-poor settings, and offers the possibility of long-term storage for future outbreaks. In summary, the robustness, low cost and ease of production of monobodies and nanobodies make them attractive candidates as potential therapeutics for the ongoing COVID-19 pandemic.
